Article Text
Abstract
Background Since the emergence of COVID-19 in December 2019, multidisciplinary research teams have wrestled with how best to control the pandemic in light of its considerable physical, psychological and economic damage. Mass testing has been advocated as a potential remedy; however, mass testing using physical tests is a costly and hard-to-scale solution.
Methods This study demonstrates the feasibility of an alternative form of COVID-19 detection, harnessing digital technology through the use of audio biomarkers and deep learning. Specifically, we show that a deep neural network based model can be trained to detect symptomatic and asymptomatic COVID-19 cases using breath and cough audio recordings.
Results Our model, a custom convolutional neural network, demonstrates strong empirical performance on a data set consisting of 355 crowdsourced participants, achieving an area under the curve of the receiver operating characteristics of 0.846 on the task of COVID-19 classification.
Conclusion This study offers a proof of concept for diagnosing COVID-19 using cough and breath audio signals and motivates a comprehensive follow-up research study on a wider data sample, given the evident advantages of a low-cost, highly scalable digital COVID-19 diagnostic tool.
- COVID-19
- virus diseases
- diagnosis
Data availability statement
Data are available upon reasonable request.
Statistics from Altmetric.com
Summary box
What are the new findings?
We demonstrate the first attempt to diagnose COVID-19 using end-to-end deep learning from a crowdsourced data set of audio samples, achieving an area under the curve of the receiver operating characteristics of 0.846.
We introduce a novel modelling strategy using a custom deep neural network to diagnose COVID-19 from a joint breath and cough representation.
We release our four stratified folds for cross-parameter optimisation and validation on a standard public corpus and details on the models for reproducibility and future reference.
How might it impact on healthcare in the future?
Our model, the COVID-19 Identification ResNet, has potential for rapid scalability, minimal cost and improving performance as more data becomes available. This could enable regular COVID-19 testing at a population scale
Introduction
COVID-19, caused by the severe-acute-respiratory-syndrome-coronavirus 2 (SARS-CoV-2), is the first global pandemic of the 21st century. Since its emergence in December 2019, it has led to over 75 million confirmed cases and more than 1.6 million deaths in over 200 countries (WHO) (https://www.who.int/emergencies/diseases/novel-coronavirus-2019). SARS-CoV-2 causes either asymptomatic infection or clinical disease, which ranges from mild to life threatening.1 Developing a swift and accurate test, able to identify both symptomatic and asymptomatic cases, is therefore essential for pandemic control.
Vocal biomarkers of SARS-CoV-2 infection have been described, thought to relate to the clinical and subclinical effects of the virus on the lower respiratory tract, neuromuscular function, senses of taste and smell and on proprioceptive feedback. Together, these produce a reduction in complexity of the coordination of respiratory and laryngeal motion in both symptomatic and asymptomatic individuals.2
We postulate that end-to-end deep learning using convolutional neural networks (CNNs) could be successfully applied to this assessment task. This article describes a proof-of-concept study of automatic symptomatic and asymptomatic COVID-19 recognition using combined breathing and coughing information from audio recordings using an end-to-end CNN design. The code for our experiments and all details for reproduction of findings can be found at https://github.com/glam-imperial/CIdeR.
Related work
Throughout the COVID-19 pandemic there have been numerous efforts to compile data sets of breath and cough audio recordings, including ‘Coughvid’,3 ‘Breathe for Science’ (https://www.breatheforscience.com), ‘Coswara’4 and ‘CoughAgainstCovid’.5 With their release, several studies have been published that leverage audio signals alongside machine learning to detect the virus.6–12 Each data set presents different challenges and so the results are often not directly comparable. Here we summarise the most relevant studies.
Brown et al6 achieved an area under the curve of the receiver operating characteristics (AUC-ROC) of 80% when classifying between COVID-positive and asymptomatic COVID-negative individuals. This was on the same data set as this study, although they did not evaluate their method on the most pertinent task of classifying all COVID-positive versus all COVID-negative cases (Task 4 in the Experiments and Results section).
On the task of classifying between COVID-positive, COVID-negative, bronchitis and pertussis cases, Imran et al12 attained 90% sensitivity from cough recordings using an ensemble of machine learning models. This was using a data set of 70 COVID-positive individuals, 226 individuals with bronchitis or pertussis and a group of 247 disease-free individuals.
Pinkas et al11 classified voice audio clips through a three-stage process of embedding the audio clips with pretrained transformers, passing these through a recurrent neural network and classifying the outputs using a support vector machine (SVM). This model had 78% sensitivity when detecting COVID-positive participants. The data set comprised 29 COVID-positive and 59 COVID-negative individuals, with recordings taken on the participants’ phones.
In a well-publicised study, Laguarta et al10 achieved 96% sensitivity on a data set of 5320 individuals, of which 2660 reportedly had COVID-19. They initialised a CNN-based model using transfer learning from other larger medical audio data sets and trained it to detect COVID-19 from extracted audio features. They achieved impressive results (notably including a suspiciously high 100% sensitivity for asymptomatic COVID-positive detection). Their results and data set have since been questioned as many of the COVID-positive participants in the data set were diagnosed based on participants’ ‘self- assessment’ of whether they had the disease.
We note that ref 10–12 have not made their code or data set publicly available, limiting the scope for reproducibility. We emphasise the importance of reproducibility, so our code and data set splits are released alongside this article.
Methods
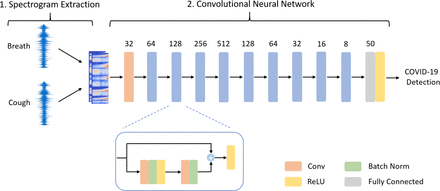
The objective is supervised learning binary classification for diagnosing COVID-19 as positive or negative using audio signals. Our implementation, displayed in figure 1, has two distinct stages which are outlined below:
1. Spectrogram extraction. As shown in figure 1, each participant in the study carried out by the University of Cambridge6 could submit waveform audio (WAV) files including a breath sample and a cough sample (Please see below and at https://www.covid-19-sounds.org/en/app/ for further details). We first compute the spectrogram of each of these WAV files to obtain a visual representation of the spectrum of audio frequencies against time. Next, we perform a log transformation, converting the spectrogram from an amplitude representation to a decibel representation. These transformations are implemented using the librosa13 python package.
A schematic of the COVID-19 Identification ResNet (CIdeR). The figure shows a blow-up of a residual block, consisting of convolutional, batch normalisation and rectified linear unit (ReLU) layers.
Each WAV file lasts between 1 and 48 s with a mean of 10 s. As uniform duration is required for CNN input, we chunk the whole WAV file into s-second segments using right padding for files shorter than s-seconds. This creates an image of size {F,W}, where F ∝ fftn and W ∝ sr ∗ s and fftn and sr are parameters used when computing the spectrogram. During model training, we only process one WAV segment (sampled uniformly). At inference time, we perform majority voting, whereby each chunk is processed in parallel, and the output label becomes the modal classification from each of the chunks (The mean of the output logits is taken in the case of a tied vote).
2. CNN. COVID-19 Identification ResNet (CIdeR) is based on ResNets,14 a variant of the CNN architecture, which uses residual blocks. As shown in figure 1, a residual block consists of two convolutions, batch normalisation15 and a rectified linear unit non-linearity. These blocks use ‘skip’ connections which add the output from these operations to the input activations for this layer. This alleviates the vanishing gradient problem, facilitating deeper architectures with more layers, thereby permitting richer hierarchical learnt representations. The number of convolutional channels for each of CIdeR’s nine layers is annotated in figure 1.
We concatenate the log spectrograms of the breath and cough samples depth-wise, creating an {F,W,2} tensor as the model input. The CNN outputs a single logit which is then passed through a sigmoid layer to obtain a (0,1) score, representing the probability of a COVID-positive sample. A weighted binary cross-entropy loss function16 is used during training to address the class imbalance in the data set.
Training strategy
Prior work6 used ‘10-fold-like’ cross-validation during training (see the paper for details). In contrast, we implement a stratified threefold cross-optimisation and additional validation partitioning using 2/1 (rotating development+train)/1 (always held-out fixed test) folds, respectively. This is to best optimise parameters independently of the test set with a small data set while ensuring that the test set remains (A) fixed for easier comparison with other work, and (B) truly blind, eliminating the possibility of CIdeR overfitting to the test set. Our stratified sampling methodology ensures that our folds represent disjoint sets of participants and each of the strata (next section) is approximately uniformly distributed across each fold. To enable reproducibility, the folds are fully released in the accompanying code.
Baseline
Our approach is not directly comparable with the study from Brown et al6 as they do not explicitly provide their folds and discard some audio samples. To this purpose, to create a performance reference for CIdeR, we implement a linear kernel SVM17 baseline. We extract openSMILE features18 for each WAV file following the Interspeech 2016 ComParE challenge format19 and perform principal component analysis,20 selecting the top 100 components by highest explained variance. We follow the cross-optimisation procedure outlined above using the development set to optimise the complexity parameter (Values between 1e−5 and 1 on a logarithmic scale) and reporting final results using the held-out test set.
Data set
The data set consists of 517 crowdsourced coughing and breathing audio recordings from 355 participants. Of these, 62 had tested positive for COVID-19 within 14 days of the recording (The data set used in this study is a small subset of the full data set that has been collected by the University of Cambridge, which has yet to be made fully public. As of July 2020, the full data set totalled 30 000 samples from roughly 16 000 participants). The samples were collected via Android and web apps developed by Brown et al6 and can be found at https://www.covid-19-sounds.org. To be classified as COVID negative, participants had to meet several stringent criteria described in ref 6.
The COVID-negative participants were divided into three categories: those without a cough (healthy-no-symptoms), those with a cough (healthy-with-cough) and those who had asthma (asthma-with-cough). The COVID-positive class consists of the 62 COVID-positive participants. This is further divided into the subclasses COVID-no-cough and COVID-cough, representing 39 COVID-positive participants without a cough and 23 participants with a cough, respectively.
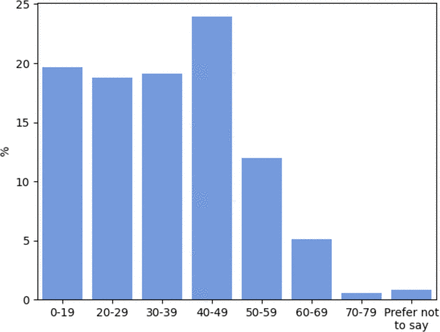
Regarding the demographics of the data set, the gender split was 38.5% female and 61.0% male (The remaining 0.5% selected Other or Prefer Not to Say). The distribution over participants’ ages is presented in figure 2. Note the positive skew and relative absence of older participants. Data regarding participants’ nationalities were also recorded and indicate that participants originate from a range of locations around the globe, including the UK, Greece, Iran and the USA. However, these data were only recorded for approximately 30% of the data set as most participants opted out of providing their geographical location, hence we do not present the international demographics here.
Participant age distribution.
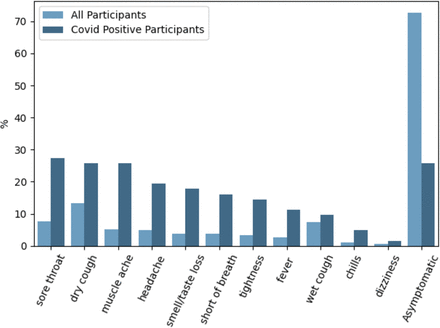
Figure 3 shows the prevalence of particular symptoms (if any) participants were experiencing at the time of audio recording. A higher proportion of the COVID-positive group were experiencing some form of symptom compared with the COVID-negative group, most commonly a sore throat. Notably, however, the second most common symptoms for COVID-positive participants were a dry cough and muscle ache, and the frequency of these two tied with that for being asymptomatic. More than half of the COVID-negative group were asymptomatic, and their most common symptom was a dry cough.
Symptoms selected by participants when providing audio recordings. All provided options to the participants are displayed in the plot. Note that the bar plots do not add up to 100%. This is due to participants being able to select more than one symptom.
Experiments and results
As indicated in the Methods section, we perform a threefold cross-optimisation using the rotating development plus train folds. Recall that the test set is fixed and always held out during optimisation. For evaluation metrics, we use the AUC-ROC and unweighted average recall (UAR), both of which are robust to imbalanced data sets. AUC-ROC maps the relationship between sensitivity and the false-positive rate as the classification threshold is varied, and UAR computes the mean recall per class. The models’ performance is sensitive to initialisation parameters, so we report the mean and SD from three training runs. Table 1 details our hyperparameter search and optimal values used for the final model.
Overview of the hyperparameter search detailing the interval, step size and optimal parameters (used to obtain the reported figures in this article—for details, see the above named GitHub repository). Hyperparameters were optimised for task 4 and subsequently used on all tasks. Adam32 was used for optimisation.
Our model performs the three tasks described in the data set publication,6 and an additional fourth task. Tasks 1–4 are as follows:
Task 1. Distinguishing between COVID positive and the stratum healthy-no-symptoms (62 vs 245 participants).
Task 2. Distinguishing between COVID-positive participants with a cough (COVID-cough) and the stratum healthy-with-cough (23 vs 30 participants).
Task 3. Distinguishing between COVID-positive participants with a cough (COVID-cough) and the stratum asthma-with-cough (23 vs 19 participants).
Task 4. Distinguishing between COVID positive and COVID negative (62 vs 293 participants).
Note that the number of participants deviates from ref 6, as we also use those audio clips shorter than 2 s resulting in partially more participants considered.
Results obtained for each task are shown in table 2, alongside the baseline. CIdeR outperforms the baseline on all tasks bar task 2 with high margin on both metrics. The results for tasks 1, 3 and 4 are statistically significant with a level of significance of 0.05 in a two-sided, two-sample t-test for difference in sample means.
Results of the models on tasks 1–4 for threefold optimisation of the number of training epochs based on the rotated development sets using the frozen optimal model parameters from table 1. Train+development/test sample counts are displayed alongside the task. Testing is performed on the held-out test fold, each. The mean area under the curve of the receiver operating characteristics curve (AUC-ROC) and the unweighted average recall (UAR) are displayed. A 95% CI is also shown following ref 33 and the normal approximation method for AUC-ROC and UAR, respectively. Scores in bold indicate significant results with α=0.05 using a two-sample t-test for no difference in means between the baseline and CIdeR based on the SD from the 3-threefold cross-optimisation.
Discussion
The results in table 2 demonstrate two key points: (1) diagnosis of COVID-19 using a CNN-based model trained on crowdsourced data is possible; (2) CIdeR obtained a high AUC-ROC of 0.846 on task 4, the task using the entire sample and hence represents the most pertinent task (symptomatic and asymptomatic COVID positive vs COVID negative). These results support the suggestion that jointly processing breath and cough audio signals using a CNN-based classifier could act as an effective and scalable method for COVID-19 detection.
The only task where CIdeR failed to outperform the baseline in our experiments was task 2. We posit this is jointly due to the small number of samples and the similarity of audio patterns between healthy participants with a cough and those with COVID-19, creating a challenging task. It is interesting that CIdeR was better able to distinguish the 19 participants with asthma and a cough from the 23 who were COVID positive with a cough (AUC-ROC 0.909). We leave further analysis for future work.
Limitations
A key limitation of this study is the size of the data set.6 We are limited to 62 COVID-positive participants and as detailed in table 2, this results in relatively wide confidence intervals for the reported metrics. One should also consider the data set demographics when analysing our results. As detailed in the Data set section, there is an under-representation of older participants, particularly 70+.
Importantly, our cohort of COVID-free participants is not a random sample, as the COVID-negative criteria required that the subject lived in a country with low COVID-19 rates, among other requirements. Hence, COVID-19 status and nationality/country of residence are correlated in the data set. This makes interpreting our results more challenging as it creates an opportunity for bias in the model. For example, we cannot conclude that CIdeR is detecting true COVID-19 features in the audio samples as opposed to, say, differences in the acoustic characteristics of breathing and coughing across nationalities, which could act as a proxy for COVID-19 status given the correlation between nationality/country of residence in the data set.
We can conclude that CIdeR has identified audio biomarkers which enable it to distinguish between the COVID-positive and COVID-negative cases for the provided data. Whether these representations would be as useful for COVID-19 status classification in the general public is still an open question. Before such a technology could be deployed, collection and evaluation on a larger, more representative data set is necessary. Conversely, since deep learning-based methods benefit from larger data set sizes, it is possible that CIdeR’s diagnostic capability could improve when used on a larger data set.
Particular attention should also be paid to ensuring that the ground truth of the data set is as close to the true classification as possible. This means demanding a reverse transcription PCR test result for the categorisation into COVID positive or COVID negative, as other forms of testing such as the lateral flow test (LFT) have high error rates.21 We note that this standard of requiring a PCR test for both positive and negative categorisations has yet to be fully adopted by the machine learning community. Some prominent studies such as Laguarta et al’s10 study have accepted ‘self-assessment’ as a ground truth diagnostic method. The time frame between the audio recording and the PCR test being conducted should also be reduced to the lowest practical period. Progress on these points is being made, with Bartl-Pokorny et al8 now collecting audio recordings at test centres. PCR tests also provide additional information to COVID-19 classification, crucially the cycle threshold which is correlated with viral load.22 Knowing the viral load of the patients at the time of recording would allow for the artificial intelligence (AI) models’ sensitivity to the level of virus to be determined, an important metric to consider when assessing the effectiveness of a mass testing scheme.23 24
Importance of asymptomatic diagnosis
While one machine learning-based approach25 has achieved an AUC-ROC of 0.862 based on demographics and symptomatology alone, this model evidently could not detect asymptomatic and presymptomatic infections. In contrast, we demonstrate that CIdeR has the potential to detect asymptomatic and presymptomatic cases, a desirable feature for COVID-19 diagnostics as a substantial proportion of virus transmission is understood to occur during the presymptomatic phase.26 Recall from figure 3 that over 25% of the COVID-positive participants in our data set were asymptomatic.
Mass testing considerations
It is also important to acknowledge the current lack of empirical evidence that mass testing (testing all members of the public independent of whether they have symptoms or not) is of benefit, with many arguing that it could have a negative effect.27 28 The antiscreening arguments are that mass testing would divert valuable resources from other more effective schemes to combat the disease and that the diagnostic accuracy of mass testing kits has not yet been adequately established.21 For example, the LFT has a poor sensitivity of 58%29 when the test is conducted by a member of the public. Particular harm can arise from false-negative results causing falsely reassured, disease-carrying individuals to reintegrate into society and propagate the disease further, perhaps to a greater degree than if they had not taken the test.
One advantage digital tests have over physical test systems is that they require relatively minimal resources and therefore would have a less adverse effect on resources available for key services. In addition, this proof-of-concept study suggests that the diagnostic accuracy could exceed that of conventional mass testing systems such as the LFT. Additionally, as the data set was collected via crowdsourcing, CIdeR should not experience a corresponding decline in performance as seen in the LFT as a result of the public taking the tests rather than medical professionals.29
While a minimal-cost method for testing for a disease seems a very attractive possibility, the effectiveness of a screening programme does not solely depend on the testing method.30 Testing the general public without the educational material and financial support to allow subjects to act appropriately following a positive test result would yield little benefit.
As alluded to in ref 31, pandemics have historically driven innovation in healthcare. But for AI-driven screening to be one of these breakthroughs from the COVID-19 pandemic, a more comprehensive data set and further research is required.
Conclusion
Wholesale testing of the population is a promising avenue for identifying and controlling the spread of COVID-19. A digital audio collection and diagnostic system could be deployed to the majority of the population and performed daily at a minimal cost, for example, as preselection for more reliable diagnoses or monitoring of spread. This study introduces the CIdeR, which demonstrates a strong proof of concept for applying end-to-end deep learning to jointly learn representations from breath and cough audio samples. Access to a larger, more representative data set could potentially enable further learning for this model and therefore, it is plausible that CIdeR’s diagnostic capabilities could significantly increase. This potential will be explored in future research.
Data availability statement
Data are available upon reasonable request.
Ethics statements
Patient consent for publication
Ethics approval
Ethics Committee of the Department of Computer Science and Technology at the University of Cambridge.
Acknowledgments
The authors give their thanks to the help provided by their colleagues Mina A Nessiem and Mostafa M Mohamed.
Footnotes
HC and AG are joint first authors.
HC and AG contributed equally.
Contributors HC and AG designed and evaluated the CIdeR. BS conceived and supervised the project. LJ contributed to the literature search, manuscript preparation and editing. PT consulted the first authors for the entirety of the project and assisted with the write-up. AB provided advice on the baseline model and helped edit the manuscript.
Funding EPSRC Center for Doctoral Training in High Performance Embedded and Distributed Systems (HiPEDS) (EP/L016796/1). DFG under Agent-based Unsupervised Deep Interactive 0-shot-learning Networks Optimising Machines’ Ontological Understanding of Sound (AUDI0NOMOUS), Reinhart Koselleck Project (442218748). Imperial College London Teaching Scholarship. UK Research and Innovation Centre for Doctoral Training in Safe and Trusted Artificial Intelligence (www.safeandtrustedai.org) (EP/S023356/1).
Disclaimer The University of Cambridge does not bear any responsibility for the analysis or interpretation of the data used herein, which represents the own view of the authors of this communication.
Competing interests None declared.
Patient and public involvement Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Provenance and peer review Not commissioned; externally peer reviewed.