Article Text
Statistics from Altmetric.com
Summary box
What are the new findings?
Our study provides a unique comparison of continuous vital sign data derived from low-cost wearable devices with those derived from conventional bedside intensive care unit monitors in a resource-limited setting.
While we encountered several challenges in the use of wearables, continuous photoplethysmography and ECG data were obtained from wearable devices in a cohort of critically ill patients with tetanus.
Heart rate variability parameters derived from wearable data mostly correlated well with those derived from bedside monitoring data.
How might it impact on healthcare in the future?
The ability to record vital sign data using simple and low-cost equipment could be a solution to the challenge of monitoring critically ill patients in resource-limited settings.
Wearable devices could provide continuous physiological data suitable for training of machine learning systems, enabling improved risk prediction thereby facilitating improved patient outcomes.
Introduction
Attentive vital sign monitoring is a key component in caring for critically ill patients. Rapid identification of physiological derangement and timely action are related to improved outcome.1 In high-income countries, this is achieved using sophisticated continuous monitoring systems facilitated by high nurse to patient ratios. In low-income and middle-income countries (LMIC), lack of staff and equipment means this is difficult to achieve.2 3 An alternative solution for resource-limited settings, enabled by recent advances in sensor technologies, is the use of low-cost wearable devices.4 Some of these devices have the additional advantage of being able to record continuous data, which allow more complex analysis and may facilitate even better risk prediction.5–7
There is, however, limited use of these devices in the unique and challenging environments of LMIC intensive care units (ICUs).3 The majority of validatory data concerning wearables come from community settings in relatively healthy ambulatory individuals.8–10 In hospital environments, the accuracy of data derived from unstable or critically ill patients is less certain.10 Studies indicate a reasonable correlation of wearable-derived heart rate measurements with nurses’ manual observations, but less when comparing respiratory rate measurements.11 12 Similarly, medical-grade wearable patches in surgical patients showed good agreement in heart rate but not respiratory rate measurements when compared with ‘gold standard’ ICU monitors.10 13 The suitability of low-cost wearables to record continuous waveform data in critically ill patients is even less certain, as these systems may be limited by high levels of noise, movement artefact and missing data.7 8 14 15
In view of this, we aimed to pilot low-cost wearable devices for continuous vital sign monitoring in critically ill patients with tetanus in a Vietnamese ICU, using two medical-grade devices able to export continuous waveform data: a patch ECG and a wrist-worn pulse oximeter. We specifically selected patients with tetanus as they have significant cardiovascular system disturbance as well as muscle spasms and increased sweating, thus posing a significant challenge for wearable monitors.16 To evaluate quality of wearable-recorded data, we computed and compared heart rate variability (HRV) data with that from state-of-the-art clinical monitors.
Methods
The wearable devices used were ePatch ECG patch monitor (ePatch V.1.0, BioTelemetry, USA) and SmartCare wrist-worn photoplethysmograph (PPG) pulse oximeter (SmartCare Analytics, UK). The ePatch records single-channel ECG output which is stored in the device and exported at the end of the recording period. The SmartCare uses a fingertip PPG sensor to derive heart rate and oxygen saturation (SpO2), which are displayed in real time on an attached wrist-worn monitor. Heart rate, SpO2 and PPG waveform data can also be transferred in real time from the SmartCare device to a receiving device (either a tablet or a smartphone) via Bluetooth.
Patients included in this report were enrolled as part of a larger study collecting vital sign monitoring data from adults with tetanus ≥16 years old admitted to the ICU at the Hospital for Tropical Diseases, Ho Chi Minh City. Patients were enrolled within 48 hours of admission to ICU and vital sign data were recorded on enrolment and after 5 days for approximately 24 hours using bedside monitors (Philips Intellivue MX550, Philips, Germany) and/or wearable devices described above. Signals were obtained with patients in the semirecumbent position. In this report we present data of patients in whom wearable and bedside monitoring data were collected simultaneously. The ePatch records ECG at a sampling rate of 256 Hz, in two channels; the SmartCare pulse oximeter records PPG and SpO2 at 100 Hz; the Intellivue records five-lead ECG at 144 Hz, PPG 125 Hz and SpO2 reported every second. Raw data acquisition from the bedside monitor was performed with the open-source VitalSignsCapture (VSCapture, www.sourceforge.net) for Philips monitor and with software provided by the manufacturer for ePatch.
ECG signals from channel 1 of ePatch and lead II of ICU monitors were used for analysis. No formal time-stamp matching was performed between devices; however, all recordings commenced within a few minutes of each other. The first and the last 5 min of each recording were trimmed to obtain stable signals. Signal loss was defined as unrecorded amplitude (0 or NA values) or unchanged amplitude for at least 300 ms for PPG, fully noisy recording for ECG and values in range (0–70) for SpO2. Signal loss ratio was computed from usable and filtered durations in minute unit.
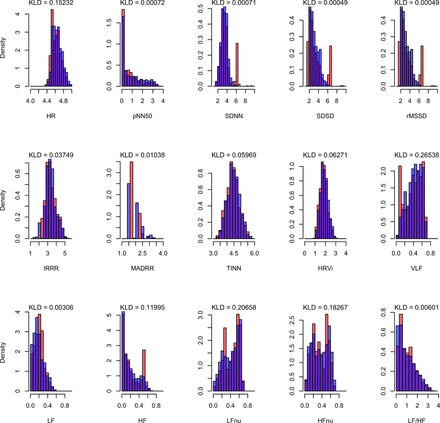
In a subset of three patients of similar disease severity, receiving similar treatment, HRV parameters were compared in 5 min segments. These parameters are measures of beat-to-beat variation in heart rate, reflecting autonomic nervous system balance. Calculation of these indices is based on mathematical evaluation of successive RR intervals and requires good-quality waveform data, and thus in calculating these we aimed to provide a clinically relevant indicator of waveform quality which could eventually be developed for disease prognostication. For this study we chose standardised measures of HRV previously found to be of value in patients with tetanus and other cardiovascular syndromes.16 17 Briefly these are divided into time domain variables (if analysed according to the distribution of RR intervals over time) and frequency domain variables if analysed after Fourier transformation (see figure 1 caption). RR intervals were identified from QRS complexes in ECG recordings using the Pan-Tompkins algorithm18 implemented in EDFBrowser (https://www.teuniz.net/edfbrowser/). For PPG, RR intervals were judged to be the intervals between systolic peaks, detected as the local maxima from preprocessed signals (slope and dominant candidate peak extraction), using the Python library scipy. The resulting RR interval series were filtered with a heart rate range of 40–200 beats per minute (bmp) and subject to HRV analysis using the R package RHRV.19 We chose to use eight time domain measures and six frequency domain measures (calculated using the Lomb-Scargle periodogram) as described in Pichot et al 17 for comparison. For each derived HRV measure, medians were tested with Wilcoxon rank-sum and the similarity between distributions tested using Kullback-Leibler divergence. To allow for multiple testing, q values are given (ie, p values adjusted for the false discovery rate).
Similarities (overlap in purple) of monitor (orange) and ePatch (blue) logarithmic heart rate variability distributions by Kullback-Leibler divergence (KLD).20 HF, high frequency; HFnu, high frequency normalised units; HR, heart rate; HRVi, integral of the density of the RR interval histogram divided by height; IRRR, IQR of RR intervals; LF, low frequency; LFnu, low frequency normalised units; LF:HF, low to high frequency ratio; MADRR, median of the absolute differences between RR intervals; pNN50, percentage of successive RR intervals that differ by more than 50 ms; rMSSD, route mean square of successive RR interval difference; SDNN, SD of NN intervals; SDSD, SD of successive RR intervals; TINN, baseline width of the RR interval histogram; VLF, very low frequency.
All subjects or their legal representatives gave written informed consent before study enrolment.
Results
Between November 2018 and June 2020, we screened 345 patients with tetanus, of whom 110 were recruited for the larger monitoring study. Due to limited availability of suitable bedside ICU monitors, there were only 19 recording days where we were able to obtain data from bedside monitors and both wearable devices (ePatch and SmartCare PPG). Additionally, there were eight recording days with data from bedside monitoring and ePatch alone, and 10 recording days with bedside monitoring and pulse oximeter data alone. Reasons for this were wearable devices falling off, faulty devices, self-removal by patients or wearable devices not being available/charged sufficiently. Of the ePatch ECG data, 7% were wholly noisy throughout the recording period due to faulty electrodes; this was not seen in bedside monitor ECG recordings. Otherwise, sporadic ECG noise sections were removable with bandpass filtering. For PPG, signal loss was approximately 1% for monitor PPG data, while only 24% of wearable data remained for analysis after the two-stage filtering described above. Remarkably, SpO2 signal loss in bedside monitor data was higher than that in wearable monitor data (38% and 26%, respectively).
Comparison of HRV features between ECG and PPG obtained by monitor and wearables is shown in table 1. There was a small difference in median frequency domain features obtained from monitors and wearable devices, whereas small but significant differences were seen in many of the time domain variables. Distributions of ECG-derived and PPG-derived HRV variables using data obtained from either monitor or wearables were similar, with Kullback-Leibler divergence mostly close to 0 (ECG data shown in figure 1; PPG data not shown). SpO2 values between monitor and wearables were similar (mean 99.08% and 99.89% for wearable and monitor, respectively).
Comparison of ECG HRV measures obtained from monitor and wearables
Discussion
Our study demonstrates the feasibility of recording high-quality continuous vital sign data from patients in an LMIC ICU using wearable devices. Both heart rate and SpO2 measurements derived from wearable monitors were similar to those derived from bedside monitors, and the order of magnitude of any differences was not of clinical significance. Our study also examined whether the data obtained from wearable devices were of sufficient quality to allow more complex analysis. We observed that particularly frequency domain parameters (indicators of spectral power) derived from these devices were similar to those from bedside ICU monitors. As time domain HRV variables should ideally be assessed from complete 24-hour recordings,20 it is possible that our comparison using short segments introduced some bias. It may also reflect the more challenging process of R peak detection with data from wearable devices where lower quality of wearable signal may have led to a worse performance in RR detection, resulting in discrepancies with the monitor ‘gold standard’.
Our study agrees with previous studies using wearable devices in that significant amounts of signal loss occurred.7 Reasons for this may be both physical (eg, poor quality or faulty devices) and contextual. Our rationale in performing this study in patients with tetanus was that they present many issues for wearable devices. For example, muscle spasms and sweating challenge the physical application of sensors, producing motion artefact and variation in cardiovascular parameters test accuracy of analytic algorithms. Additionally, contextual factors we have not formally examined, such as Wi-Fi internet and 3G/4G network, may also affect signal quality and stability.
Practically, sensor loss was a common occurrence for both patch and fingertip sensors. As all patients had bedside ICU monitors for routine monitoring, loss of additional sensors was likely to be unnoticed by nursing staff. For ePatch devices where data are stored internally during recording, this issue was further compounded as study staff could not verify data recording in real time. In view of this, we feel that real-time data visualisation and quality feedback are likely to be an important factor in maximising data quantity and quality collected from wearable devices.
In addition to these issues, perhaps the largest challenge we encountered was the limited choice of suitable devices to use in this study as we required devices from which continuous data could be exported. This applied to both ICU monitors and wearables. In future, this may be even more challenging as more device manufacturers restrict data access. In high-income settings, there are costly systems permitting access, but alternative solutions are required for LMICs. While we have found there are significant difficulties in the practical application of wearable devices in a real-world setting, advances in machine learning technology offer potential to address issues of data loss and compensate for signal quality, making wearable systems a more realistic solution. Machine learning and other artificial intelligence techniques could also be used to perform rapid and complex analysis of the complex physiological waveform data such as those recorded here. In high-income settings, these methods have been shown to better identify patients at risk of deterioration compared with standard monitoring approaches,5 and could be used similarly in our setting to improve risk prediction and patient outcomes.
Ethics statements
Ethics approval
The study was approved by both the Ethics Committee of the Hospital for Tropical Diseases and the Oxford Tropical Research Ethics Committee.
Acknowledgments
The authors would like to acknowledge the staff and patients at the Adult Intensive Care Unit of the Hospital for Tropical Diseases.
References
Footnotes
HMTV and NVH are joint first authors.
DC and CLT are joint senior authors.
Twitter @phan189
Collaborators Vietnam ICU Translational Applications Laboratory (VITAL) Investigators: OUCRU inclusive authorship list in Vietnam (alphabetic order by surname): Dang Trung Kien, Dong Huu Khanh Trinh, Joseph Donovan, Du Hong Duc, Ronald Geskus, Ho Bich Hai, Ho Quang Chanh, Ho Van Hien, Hoang Minh Tu Van, Huynh Trung Trieu, Evelyne Kestelyn, Lam Minh Yen, Le Dinh Van Khoa, Le Nguyen Thanh Nhan, Luu Phuoc An, Nguyen Lam Vuong, Nguyen Than Ha Quyen, Nguyen Thi Le Thanh, Nguyen Thi Phuong Dung, Ninh Thi Thanh Van, Phan Nguyen Quoc Khanh, Phung Khanh Lam, Phung Tran Huy Nhat, Guy Thwaites, Louise Thwaites, Tran Minh Duc, Trinh Manh Hung, Hugo Turner, Jennifer Ilo Van Nuil, Sophie Yacoub. Hospital for Tropical Diseases, Ho Chi Minh City (alphabetic order by surname): Cao Thi Tam, Duong Bich Thuy, Ha Thi Hai Duong, Ho Dang Trung Nghia, Le Buu Chau, Luong Thi Hue Tai, Nguyen Hoan Phu, Nguyen Quoc Viet, Nguyen Thanh Nguyen, Nguyen Thanh Phong, Nguyen Thi Kim Anh, Nguyen Van Hao, Nguyen Van Thanh Duoc, Nguyen Van Vinh Chau, Pham Kieu Nguyet Oanh, Phan Tu Qui, Phan Vinh Tho. University of Oxford (alphabetic order by surname): David Clifton, Mike English, Heloise Greeff, Huiqi Lu, Jacob McKnight, Chris Paton. Imperial College London
(alphabetic order by surname): Pantellis Georgiou, Bernard Hernandez Perez, Kerri Hill-Cawthorne, Alison Holmes, Stefan Karolcik, Damien Ming, Nicolas Moser, Jesus Rodriguez Manzano. King’s College London (alphabetic order by surname): Alberto Gomez, Hamideh Kerdegari, Marc Modat, Reza Razavi. ETH Zurich (alphabetic order by surname): Abhilash Guru Dutt, Walter Karlen, Michaela Verling, Elias Wicki. The University of Melbourne (alphabetic order by surname): Linda Denehy, Thomas Rollinson.
Contributors NVH, TZ, DC and CLT conceived the study. HMTV, NVH, LMY, HTHD, DBT, TZ, HG, DC and CLT designed the study. HMTV, NVH, KPNQ, HBH, LDVK, LMY, PTHN, HTHD and DBT implemented the study. HMTV, HBH, LMY and HG analysed the data. NVH, DC and CLT resourced the study. HMTV, HBH, LDVK, HG, HTHD, DC and CLT drafted the initial draft. All authors critically reviewed the final manuscript.
Funding The study was funded by the Wellcome Trust (217650/Z/19/Z). HMTV, KPNQ, HBH, LDVK, LMY, PTHN, HG and CLT were supported by the Wellcome Trust (107367/Z/15/Z, 089276/B/09/7 and 217650/Z/19/Z). DC was supported by the National Institute for Health Research (NIHR) Oxford Biomedical Research Centre (BRC).
Competing interests None declared.
Provenance and peer review Not commissioned; externally peer reviewed.